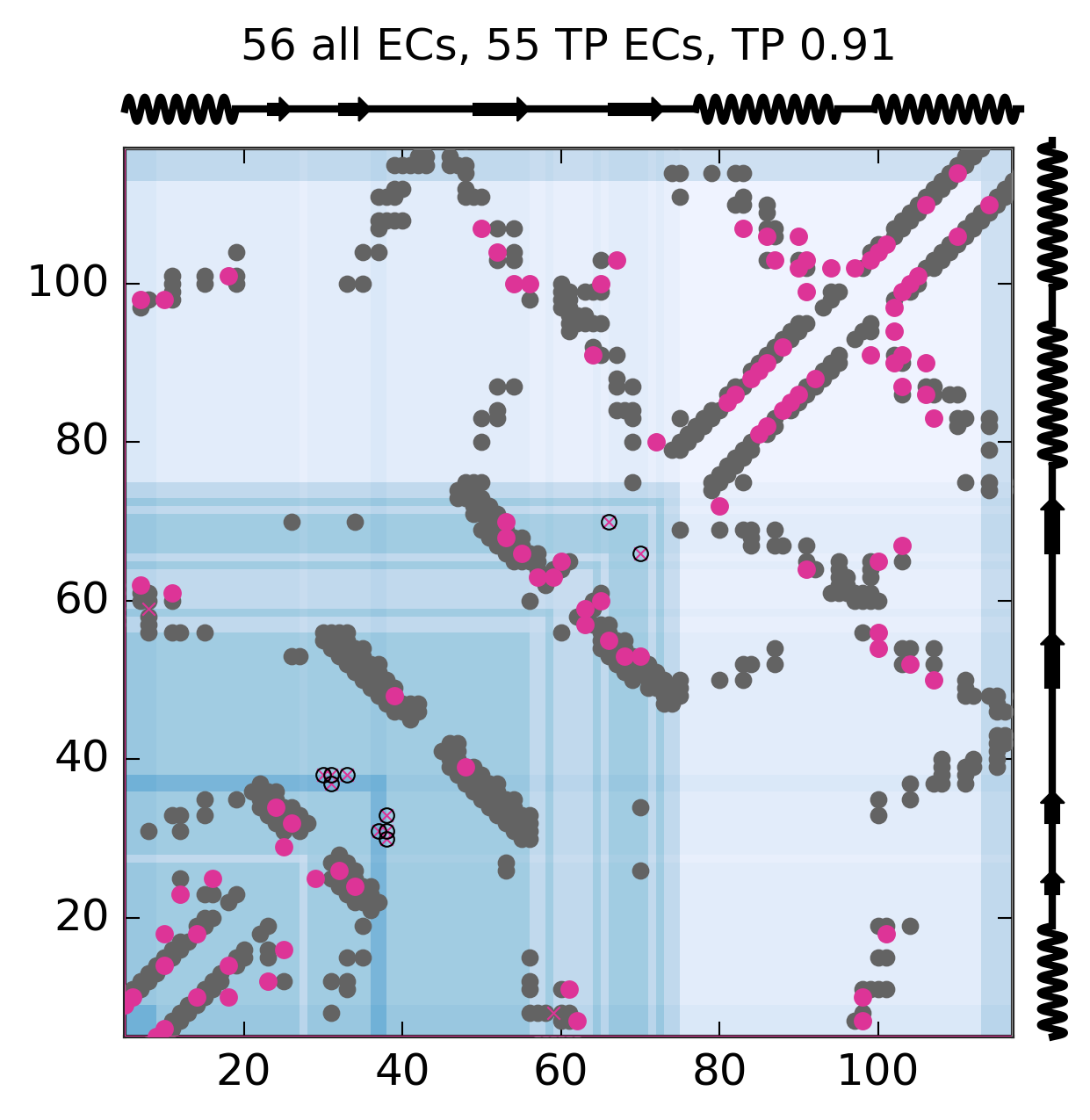
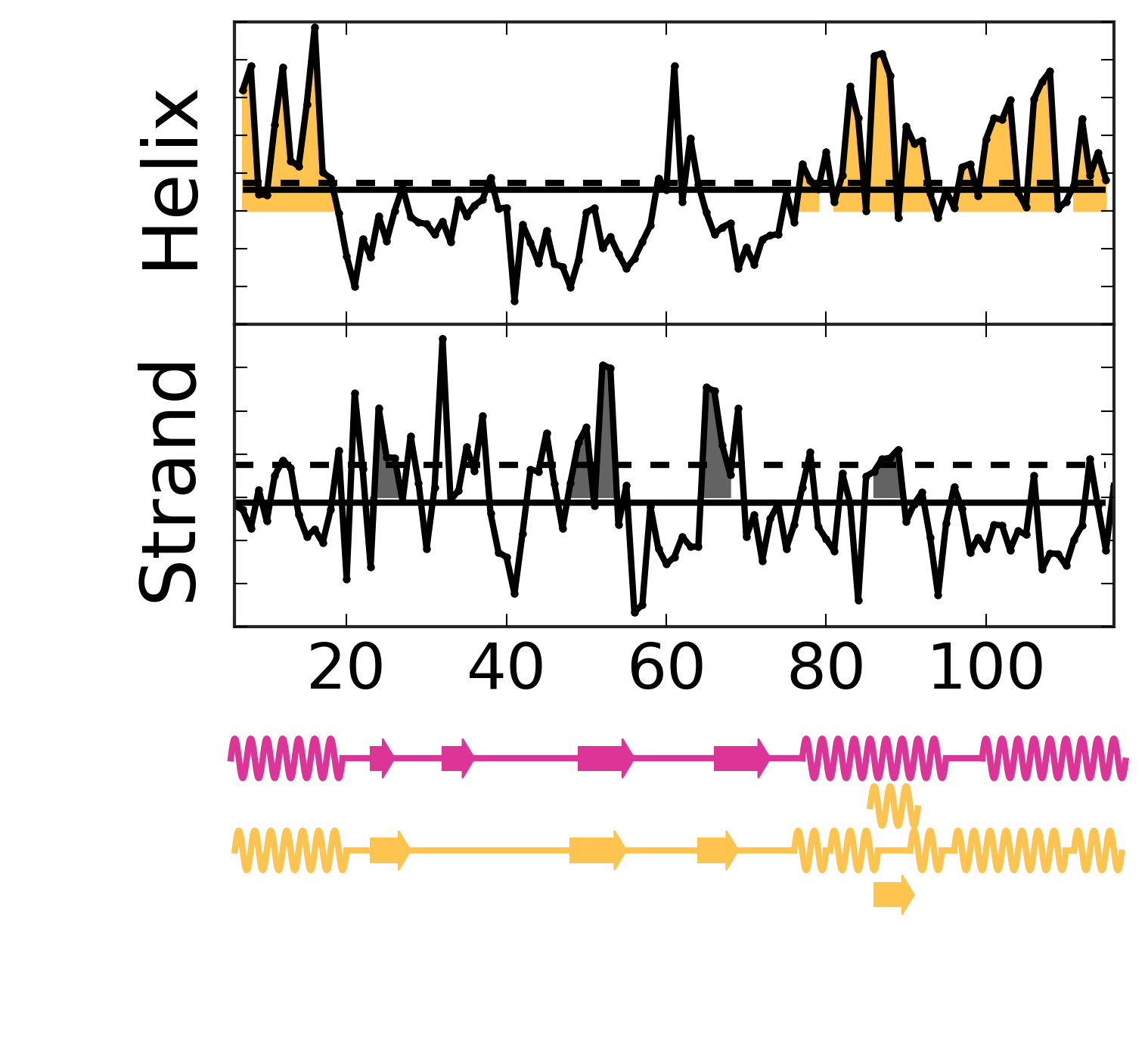
Evolutionary Coupling Analysis
Predicted and experimental contacts
Secondary structure from ECs
EC score distribution and threshold
Top ECs
| Rank |
Residue 1 |
Amino acid 1 |
Residue 2 |
Amino acid 2 |
EC score |
| 1 |
87 |
L |
103 |
L |
0.57 |
| 2 |
59 |
K |
63 |
E |
0.56 |
| 3 |
91 |
A |
103 |
L |
0.51 |
| 4 |
84 |
L |
88 |
A |
0.47 |
| 5 |
7 |
E |
98 |
V |
0.46 |
| 6 |
106 |
A |
110 |
K |
0.44 |
| 7 |
64 |
A |
91 |
A |
0.44 |
| 8 |
18 |
I |
101 |
F |
0.43 |
| 9 |
57 |
S |
63 |
E |
0.43 |
| 10 |
14 |
A |
18 |
I |
0.42 |
| 11 |
10 |
N |
14 |
A |
0.42 |
| 12 |
55 |
T |
66 |
L |
0.41 |
| 13 |
6 |
E |
10 |
N |
0.41 |
| 14 |
90 |
Q |
106 |
A |
0.39 |
| 15 |
91 |
A |
99 |
M |
0.39 |
| 16 |
56 |
Y |
100 |
I |
0.39 |
| 17 |
86 |
L |
106 |
A |
0.39 |
| 18 |
52 |
L |
104 |
V |
0.38 |
| 19 |
83 |
I |
107 |
V |
0.36 |
| 20 |
94 |
N |
102 |
T |
0.36 |
| 21 |
5 |
G |
9 |
R |
0.36 |
| 22 |
97 |
M |
102 |
T |
0.35 |
| 23 |
24 |
T |
34 |
T |
0.35 |
| 24 |
99 |
M |
103 |
L |
0.34 |
| 25 |
86 |
L |
90 |
Q |
0.33 |
| 26 |
81 |
S |
85 |
K |
0.33 |
| 27 |
110 |
K |
114 |
I |
0.32 |
| 28 |
101 |
F |
105 |
T |
0.32 |
| 29 |
85 |
K |
89 |
L |
0.32 |
| 30 |
31 |
P |
38 |
T |
0.32 |
| 31 |
88 |
A |
92 |
E |
0.31 |
| 32 |
82 |
D |
86 |
L |
0.31 |
| 33 |
53 |
K |
70 |
F |
0.29 |
| 34 |
33 |
F |
38 |
T |
0.29 |
| 35 |
100 |
I |
104 |
V |
0.29 |
| 36 |
50 |
T |
107 |
V |
0.28 |
| 37 |
60 |
Y |
65 |
P |
0.27 |
| 38 |
67 |
Y |
103 |
L |
0.26 |
| 39 |
16 |
E |
25 |
V |
0.26 |
| 40 |
12 |
L |
23 |
F |
0.26 |
| 41 |
26 |
L |
32 |
S |
0.25 |
| 42 |
72 |
Q |
80 |
V |
0.25 |
| 43 |
53 |
K |
68 |
E |
0.25 |
| 44 |
90 |
Q |
102 |
T |
0.25 |
| 45 |
54 |
F |
100 |
I |
0.25 |
| 46 |
66 |
L |
70 |
F |
0.25 |
| 47 |
65 |
P |
100 |
I |
0.25 |
| 48 |
7 |
E |
62 |
D |
0.25 |
| 49 |
39 |
S |
48 |
V |
0.25 |
| 50 |
11 |
E |
61 |
P |
0.24 |
| 51 |
30 |
P |
38 |
T |
0.24 |
| 52 |
10 |
N |
98 |
V |
0.24 |
| 53 |
10 |
N |
18 |
I |
0.24 |
| 54 |
31 |
P |
37 |
V |
0.24 |
| 55 |
25 |
V |
29 |
N |
0.24 |
| 56 |
8 |
Q |
59 |
K |
0.24 |
Alignment robustness analysis
First most common residue correlation
Second most common residue correlation